DREAMS: deep read-level error model for sequencing data applied to low-frequency variant calling and circulating tumor DNA detection, Genome Biology
Por um escritor misterioso
Last updated 13 abril 2025
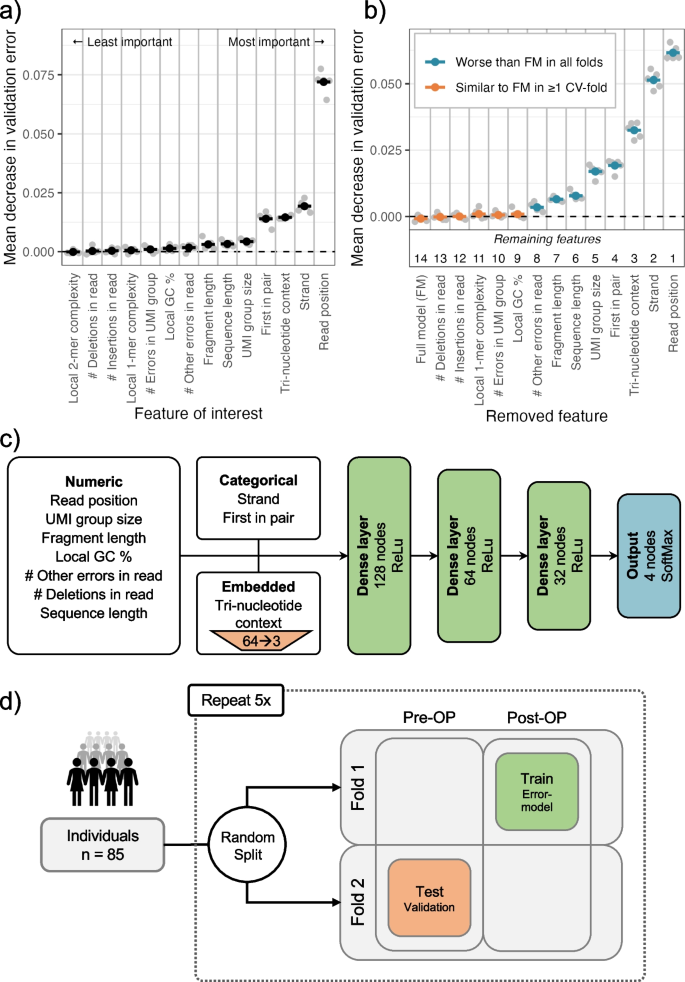
Circulating tumor DNA detection using next-generation sequencing (NGS) data of plasma DNA is promising for cancer identification and characterization. However, the tumor signal in the blood is often low and difficult to distinguish from errors. We present DREAMS (Deep Read-level Modelling of Sequencing-errors) for estimating error rates of individual read positions. Using DREAMS, we develop statistical methods for variant calling (DREAMS-vc) and cancer detection (DREAMS-cc). For evaluation, we generate deep targeted NGS data of matching tumor and plasma DNA from 85 colorectal cancer patients. The DREAMS approach performs better than state-of-the-art methods for variant calling and cancer detection.
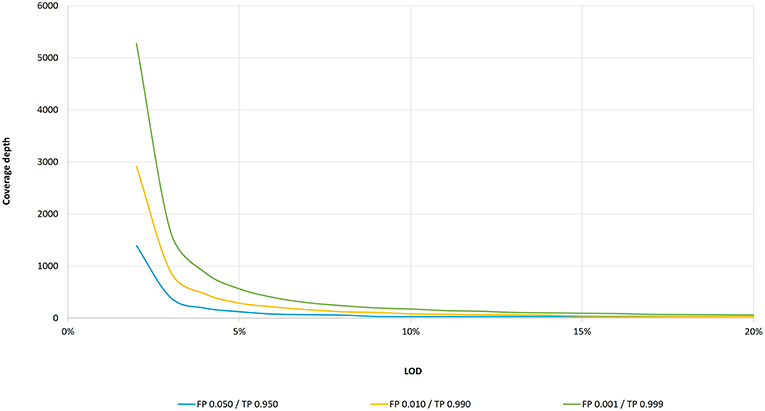
Frontiers Standardization of Sequencing Coverage Depth in NGS

PDF) DREAMS: deep read-level error model for sequencing data

Additional file 1 of DREAMS: deep read-level error model for

Sequence motifs at systematic error sites. (a) The motif around
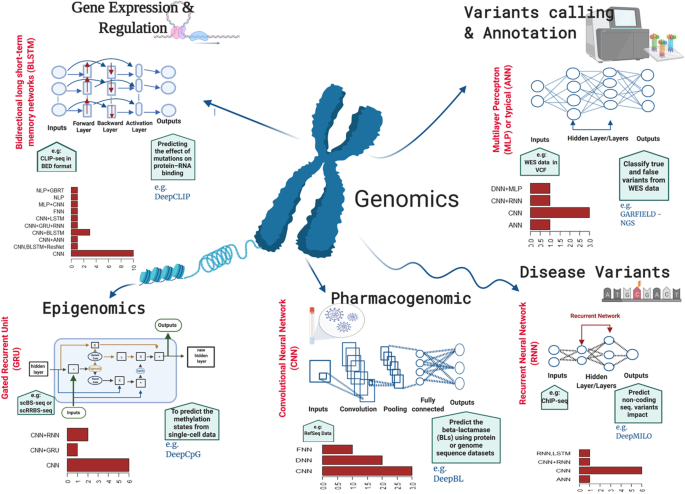
A review of deep learning applications in human genomics using

Machine learning guided signal enrichment for ultrasensitive
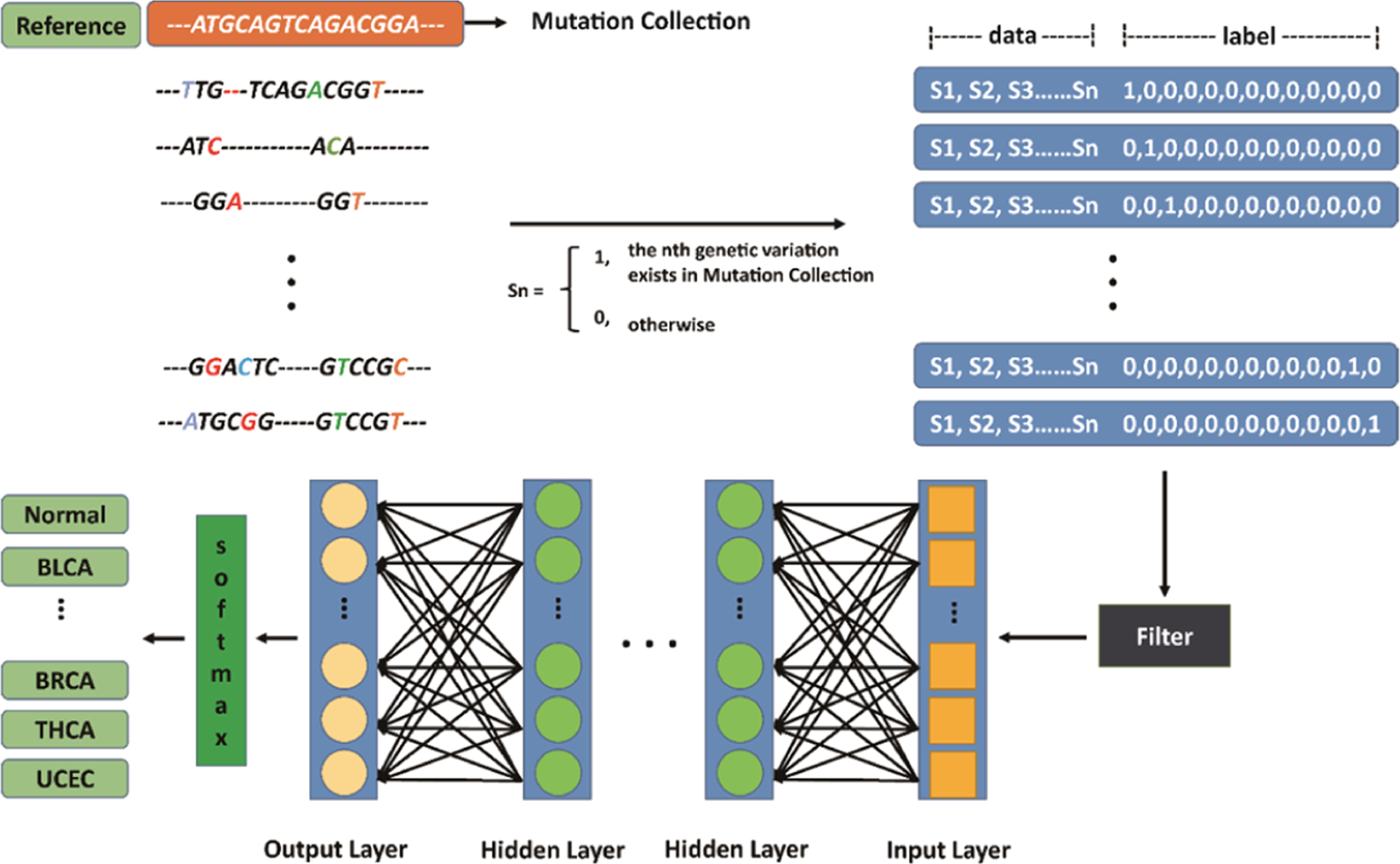
Identification of 12 cancer types through genome deep learning
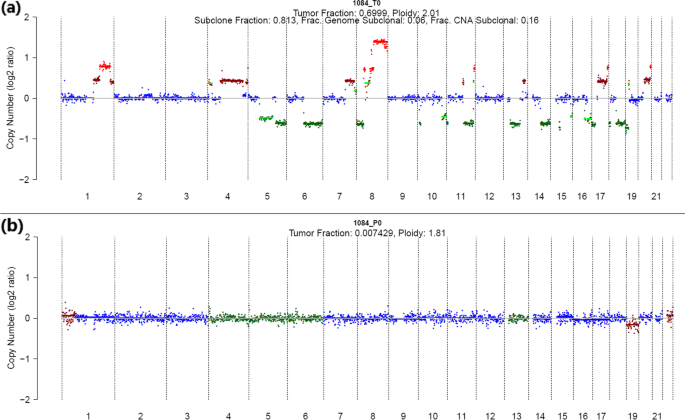
Whole genome deep sequencing analysis of cell-free DNA in samples

Integration of intra-sample contextual error modeling for improved
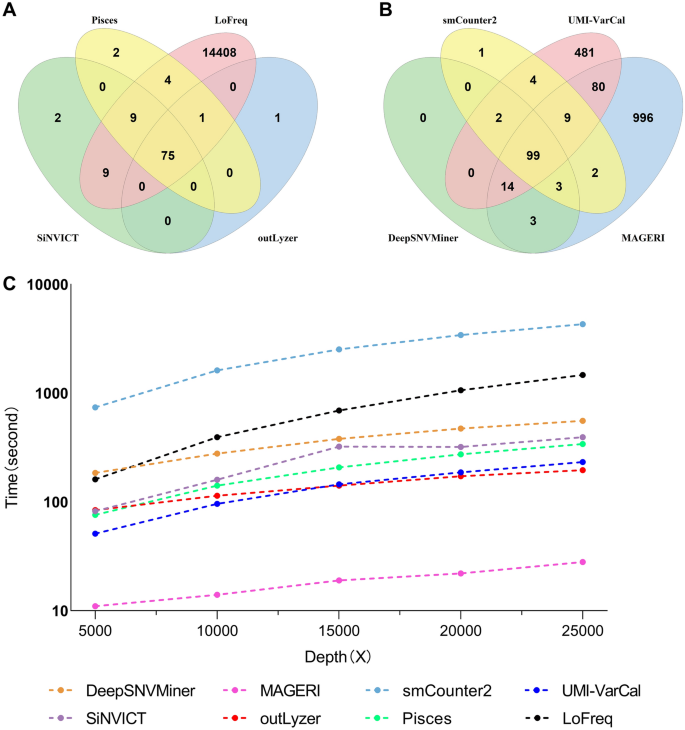
Evaluating the performance of low-frequency variant calling tools

Somatic small-variant calling methods in Illumina DRAGEN
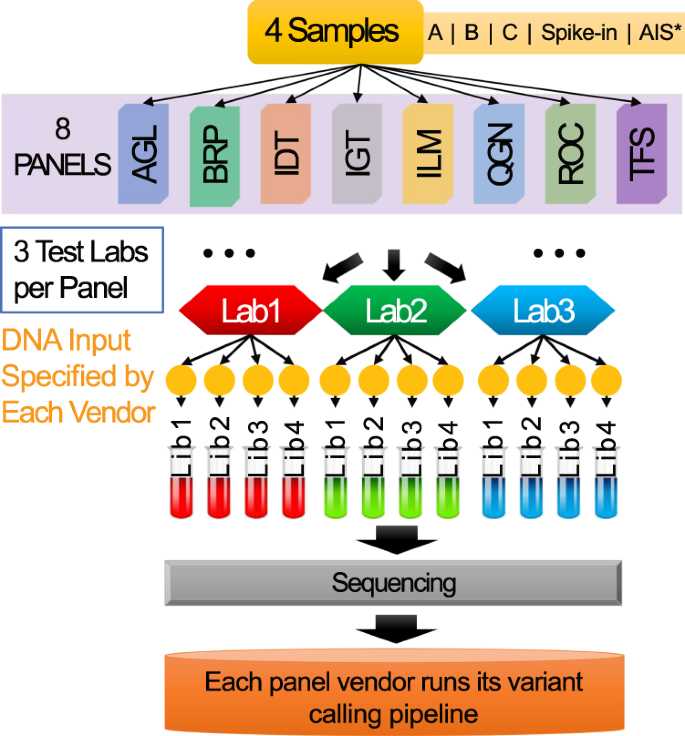
Ultra-deep multi-oncopanel sequencing of benchmarking samples with
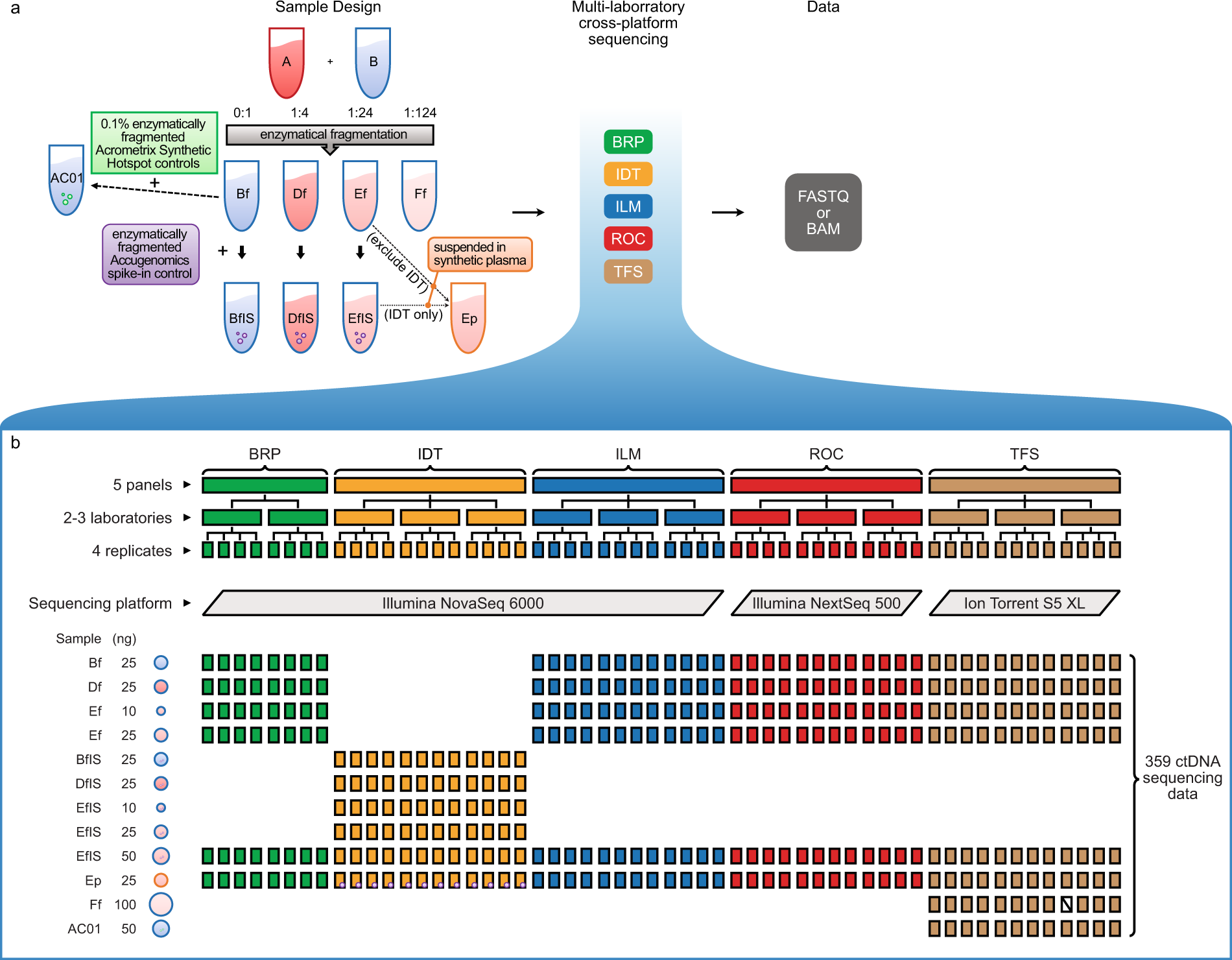
Ultra-deep sequencing data from a liquid biopsy proficiency study
Recomendado para você
-
 O P;, Object Class: N/A1 Special Containment J 7143-J being a doorknob, no 53.71.34, w,, containment procedures are líªºrkuoh Procedures: Due to SCP- Description: SCP-7143-J is a doorknob on the door to13 abril 2025
O P;, Object Class: N/A1 Special Containment J 7143-J being a doorknob, no 53.71.34, w,, containment procedures are líªºrkuoh Procedures: Due to SCP- Description: SCP-7143-J is a doorknob on the door to13 abril 2025 -
scp #scpshowerthoughts #minecraft #minecraftparkour #scp173 #scpsecre13 abril 2025
-
 press F for 7143-J : r/DankMemesFromSite1913 abril 2025
press F for 7143-J : r/DankMemesFromSite1913 abril 2025 -
 scpwiki13 abril 2025
scpwiki13 abril 2025 -
 Machimaho: I Messed Up and Made the Wrong Person Into a Magical Girl! - AnimeSuki Forum13 abril 2025
Machimaho: I Messed Up and Made the Wrong Person Into a Magical Girl! - AnimeSuki Forum13 abril 2025 -
 Cute Eye Emoji Art Board Prints for Sale13 abril 2025
Cute Eye Emoji Art Board Prints for Sale13 abril 2025 -
 Neuroanatomical organization and functional roles of PVN MC4R pathways in physiological and behavioral regulations13 abril 2025
Neuroanatomical organization and functional roles of PVN MC4R pathways in physiological and behavioral regulations13 abril 2025 -
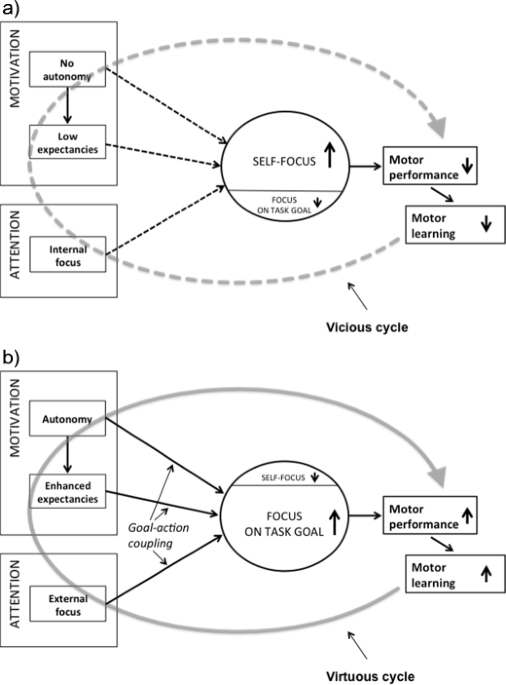 Optimizing performance through intrinsic motivation and attention for learning: The OPTIMAL theory of motor learning13 abril 2025
Optimizing performance through intrinsic motivation and attention for learning: The OPTIMAL theory of motor learning13 abril 2025 -
 Transgenic Mice Expressing Green Fluorescent Protein under the Control of the Melanocortin-4 Receptor Promoter13 abril 2025
Transgenic Mice Expressing Green Fluorescent Protein under the Control of the Melanocortin-4 Receptor Promoter13 abril 2025 -
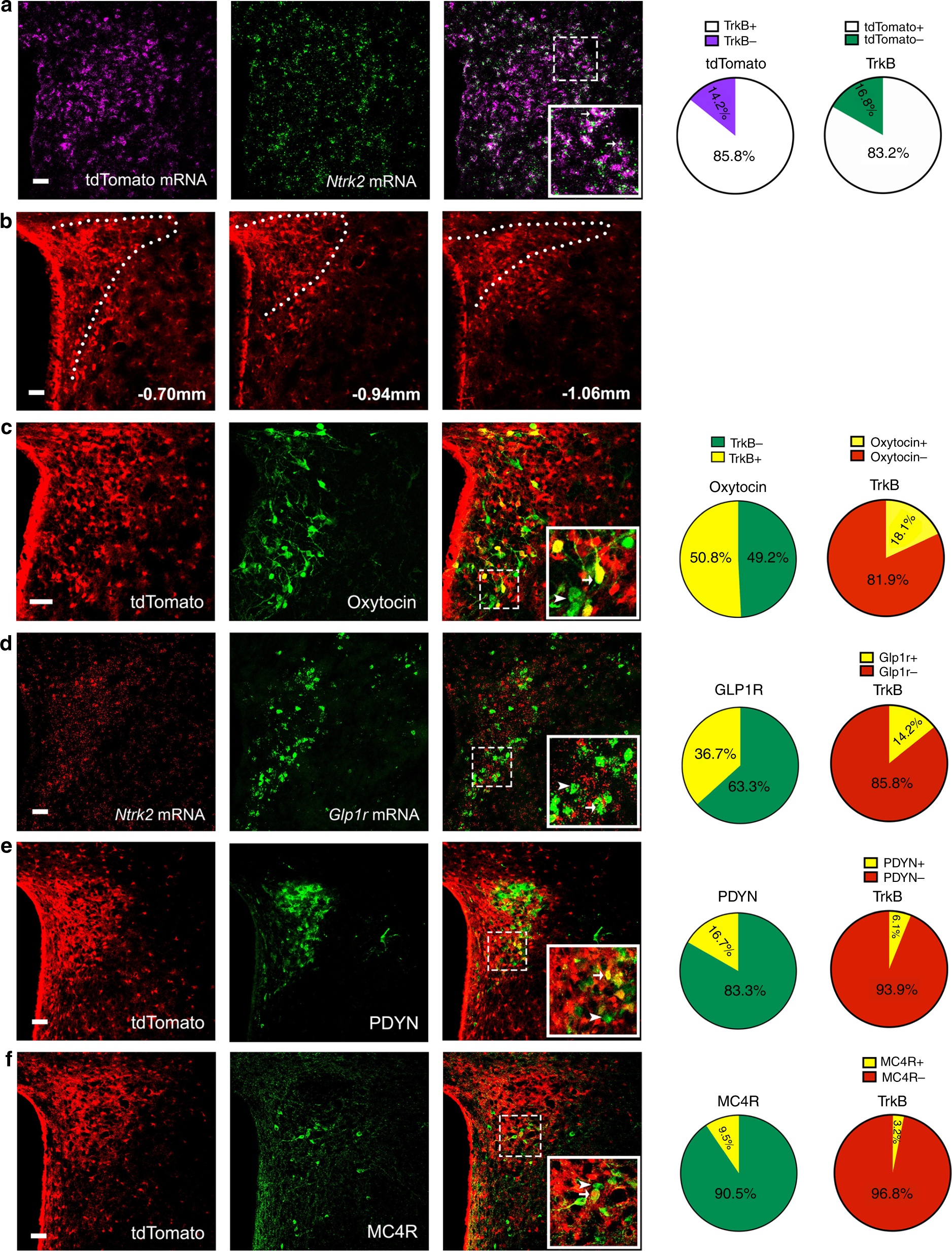 TrkB-expressing paraventricular hypothalamic neurons suppress13 abril 2025
TrkB-expressing paraventricular hypothalamic neurons suppress13 abril 2025
você pode gostar
-
 Mural 'Já não somos indefesas13 abril 2025
Mural 'Já não somos indefesas13 abril 2025 -
 Garfo para Churrasco Alumínio 4 Pontas13 abril 2025
Garfo para Churrasco Alumínio 4 Pontas13 abril 2025 -
 Perto do Verdão, Moisés posta mensagem de despedida na Croácia - Futebol - UOL Esporte13 abril 2025
Perto do Verdão, Moisés posta mensagem de despedida na Croácia - Futebol - UOL Esporte13 abril 2025 -
Demon King Daimao Sai Akuto's Imperial Capital War - Watch on13 abril 2025
-
 5 panel gru meme - Imgflip13 abril 2025
5 panel gru meme - Imgflip13 abril 2025 -
Buy Layers of Fear: Inheritance Steam Key GLOBAL - Cheap - !13 abril 2025
-
 Convite Homem Aranha Edite Online13 abril 2025
Convite Homem Aranha Edite Online13 abril 2025 -
How Do I Enable The New Samsung Assistant Sam On M - Samsung13 abril 2025
-
 Campeonato Mineiro da Segunda Divisão já sofre uma baixa13 abril 2025
Campeonato Mineiro da Segunda Divisão já sofre uma baixa13 abril 2025 -
 Z From Alphabet Lore by g4merxethan on DeviantArt13 abril 2025
Z From Alphabet Lore by g4merxethan on DeviantArt13 abril 2025


